RNA model introduction
Description of the oxRNA model
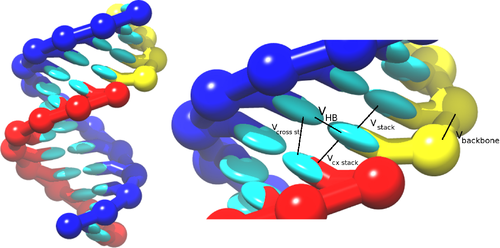
The RNA model, oxRNA, treats each RNA nucleotide as a single rigid body with multiple interaction sites, following the coarse-graining approach adopted for the DNA model.
The nucleotides interact with the following pairwise interaction potentials:
- Backbone connectivity ,
- Excluded volume ,
- Hydrogen bonding ,
- Nearest-neighbor stacking ,
- Cross-stacking in a duplex ,
- Coaxial stacking .
which are schematically illustrated in the picture:
Simulation units
The code uses units for energy, mass, length and time that are convenient for a typical system. The relationship between simulation units (SU) and SI units is given below.
| Simulation unit | Physical unit |
|---|---|
| 1 unit of length | |
| 1 unit of energy | |
| 1 unit of temperature | |
| 1 unit of force | |
| 1 unit of mass | |
| 1 unit of time |
Running a simulation with the oxRNA model
The oxRNA model is integrated into the oxDNA simulation code. In particular, it is possible to use the Virtual Move Monte Carlo (VMMC), Monte Carlo (MC) and Molecular Dynamics (MD) simulation algorithms using the same format of input file as for the DNA model. The format of the configuration files is also the same as for the DNA model, described in Documentation. When running simulations of the oxRNA model, the following additional line must be included in the input file to specify that the RNA model is to be used:
interaction_type = RNA
The RNA model comes with two parametrizations, the average-base and sequence-dependent one. In the average-base parametrization, the interaction strengths are the same for all Watson-Crick and wobble base pairs (AU, GC, GU) and 0 for all other types of base pairs, and the interaction strengths have the same strength for all possible pairs of nucleotides interacting with the stacking interaction . In the sequence-dependent version of the model, the interaction strengths of and depend on the type of interacting bases (interactions for are still 0 for base pairs other than AU, GC or GU).
The average-base parametrization is used by default. In order to use the sequence-dependent version of the model, the following options need to be added into the input file:
use_average_seq = 0 seq_dep_file = rna_sequence_dependent_parameters.txt
Note that the file rna_sequence_dependent_parameters.txt needs to be located in the directory where you run the simulation, or the full location of the file needs to be specified in the seq_dep_file option.
Furthermore, the initial configuration files need to be generated so that the nucleotides are positioned in an arrangement that satisfies the RNA potentials (for instance in the case of a duplex, they need to be initialized in an A-helical structure). For this purpose, a script generate-RNA.py is provided in the UTILS/ subdirectory of the source code main directory. For instance, if one wants to generate an initial configuration consisting of three strands, two of them complementary (with sequence 3'-GCAAGUCG-5' and its complementary) in a duplex configuration, and one single strand with sequence 3'-ACCCGU-5', one needs to create the following text file, called for example sequences.txt:
DOUBLE GCAAGUCG ACCCGU
Note that the sequences are always specified in 3'-5' order. In order to create the initial configuration files generated.top and generated.conf with the duplex and single strand randomly placed in a simulation cube with side of length 20 in simulation units, run the script
generate-RNA.py sequences.txt generated 20.0
which will create the configuration files. These can then be used as an initial configuration for a simulation. Other input file options that apply to oxDNA, such external_forces=1 (for the use of external forces), can be used with oxRNA with the same syntax (see Documentation for a full list and for further details).
For an example on how to use VMMC simulations to determine the melting temperature of an RNA duplex, please see the RNA duplex melting tutorial.
The latest version of the code also includes Debye-Huckel potential for electrostatic interactions. RNA2 version of the code needs to be used if one wants to include electrostatic effects at given salt concentration. For example, to run the code at 0.5M salt concentration, include the following in the input file:
interaction_type = RNA2 salt_concentration = 0.5
Visualization of RNA configurations
In order to visualize the configurations of the oxRNA model, one can use the traj2chimera.py script, as described for the oxDNA model. It is however necessary to first set the environment variable OXRNA to 1 in order for the script to properly generate visual representation of oxRNA:
export OXRNA=1
The visualization of a configuration specified in, for example, generated.top and generated.conf can then be obtained by running
traj2chimera.py generated.conf generated.top
in the UTILS/ directory which creates the files generated.conf.pdb and chimera.com which can then be visualized with Chimera software by running the following command:
chimera generated.conf.pdb chimera.com
or alternatively, you can load generated.conf.pdb in the Chimera software and then click on Tools->General Controls->Command line and specify
read chimera.com
in the command line, where chimera.com needs to be present in the directory where you started Chimera.
References
The model and its performance is discussed in detail in the following reference:
P. Šulc, F. Romano, T. E. Ouldridge, J. P. K. Doye, A. A. Louis: A nucleotide-level coarse-grained model of RNA, : The Journal of Chemical Physics 140, 235102 (2014)
The extension to include salt-dependent effects is described in the supplementary material of the following reference:
C. Matek, P. Šulc, F. Randisi, J.P.K. Doye, A. A. Louis: Coarse-grained modelling of supercoiled RNA J. Chem. Phys. 143, 243122 (2015)